Projects
On-the-fly (embryo) computing
PI: Angela Stathopoulos (Division of Biology and Biological Engineering)
SASE: Dave Rumph, Senior Software Engineer
Because the complete genome of many species, including humans, has now been mapped, many may be under the impression that there is little more to learn about the generics of various animals. Nothing could be further from the truth! The arena of gene expression—when different genes are active and how they are controlled—has many mysteries to unravel. For example, when a fertilized egg first starts to divide, all the cells are the same, but quickly the cells start to differentiate, ultimately forming vastly different tissues and organs such as muscle, nerves, and skin. Little is known about what directs this process.
The lab of Angela Stathopoulos is conducting research using the embryos of the fruit fly, Drosophiila melanogaster, to understand these mechanisms, particularly how "cis regulatory modules” are involved. The experimental technique relies on tagging different portions of DNA in live embryos with probes that fluoresce when illuminated with different wavelengths of laser light in a confocal microscope. The resulting set of three-dimensional images can be used to determine the spatial orientation of three different areas of the chromosome. Their relative positions indicate which portion of the chromosome are controlling the expression of a gene during each stage of embryo development.
The raw microscope images require processing to identify, group and categorize the fluorescing spots. An amalgam of scripts and code had been written in the past by graduate students and consultants, but was fragile, difficult to use and maintain, and supported only a now-obsolete microscope image format. The Schmidt Academy collaborated with the Stathopoulos lab to rewrite and update of the code in modern Python, with code that supports the new microscope format while providing a set of visual tools to allow fine-tuning the parameters involved with each step of the data processing. The new code includes an easy-to-use graphical user interface, and runs on Mac, Windows and Linux computers.
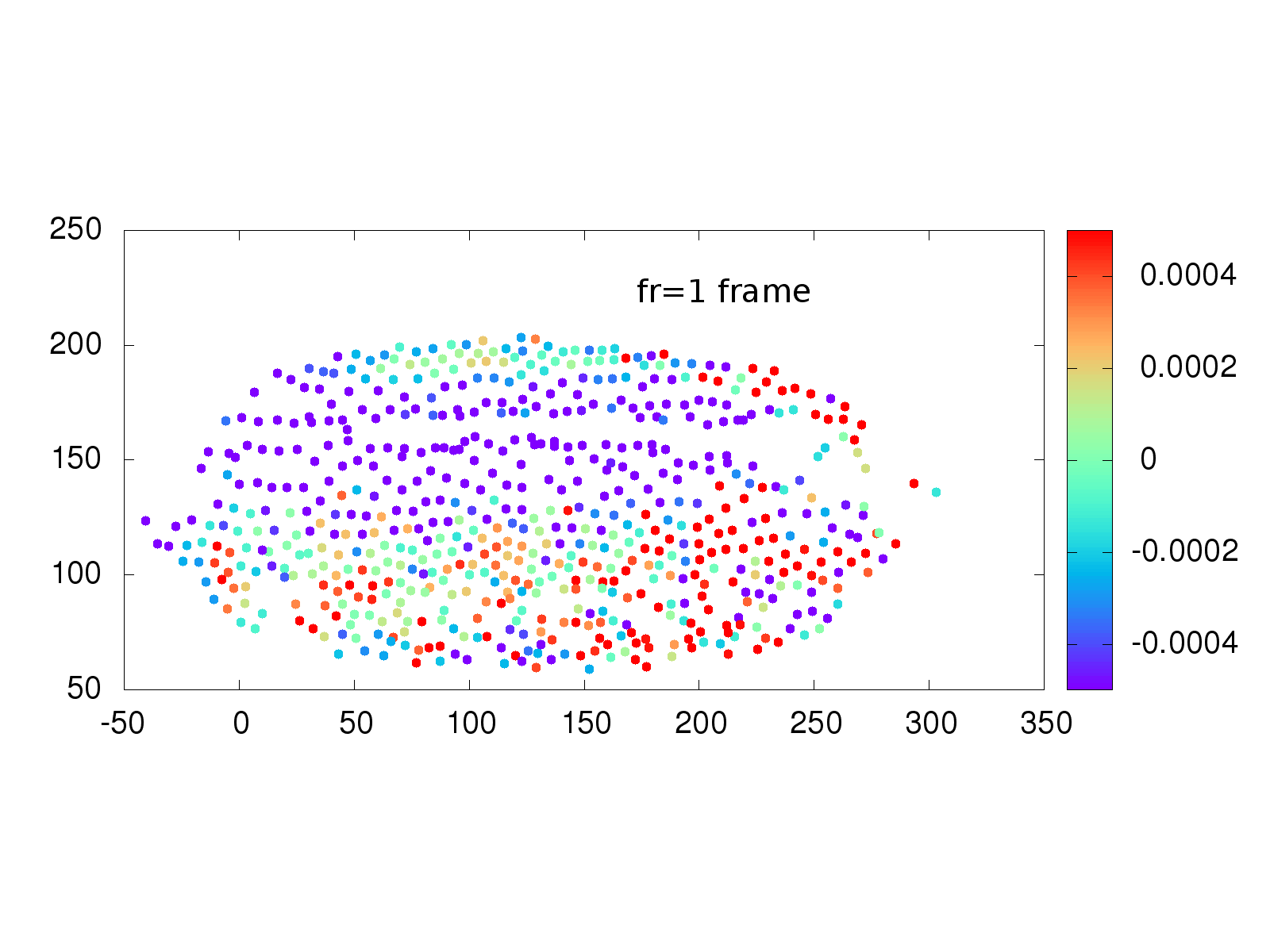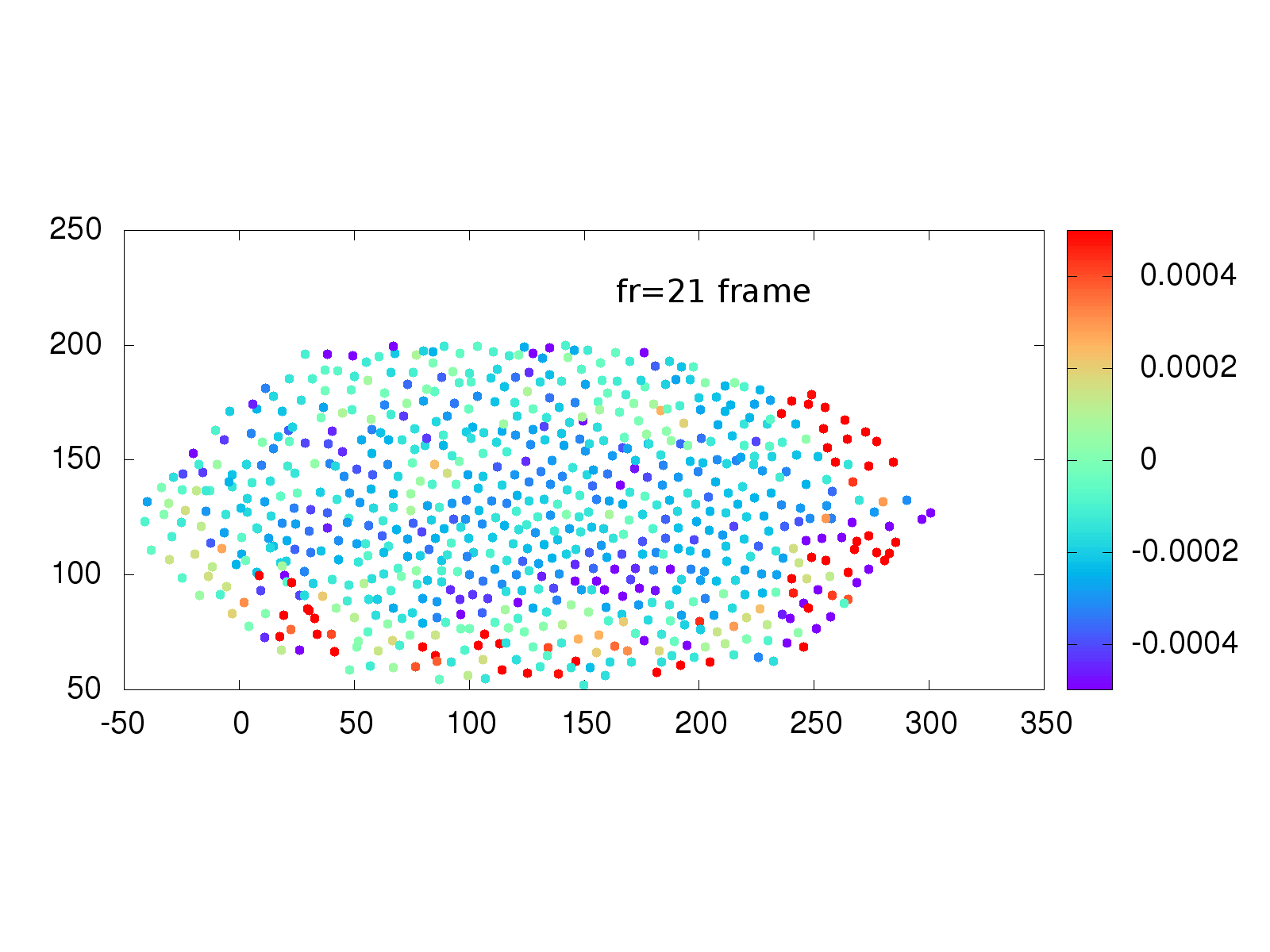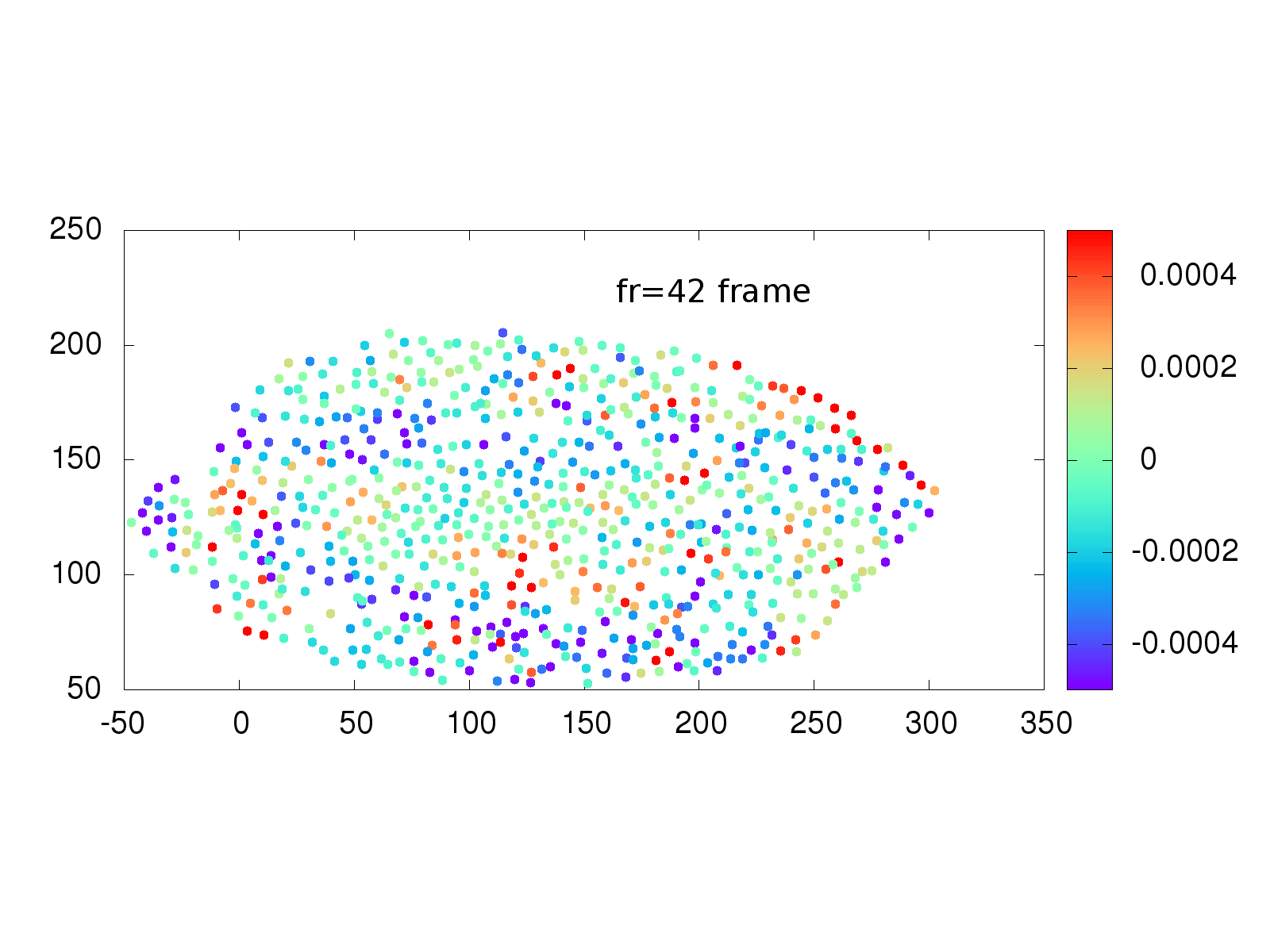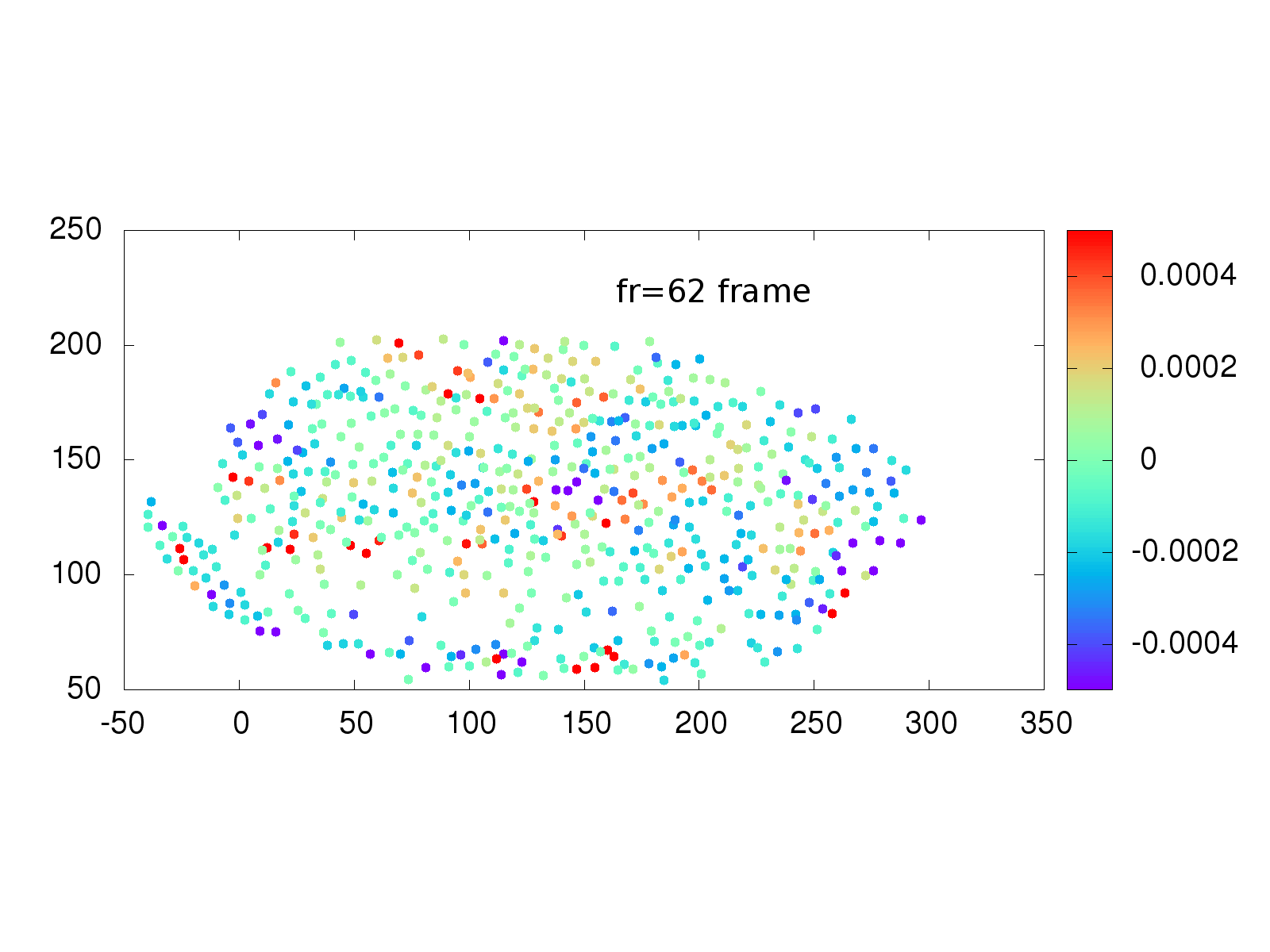
Another area of interest to the Stathopoulos lab is whether physical strain on cells from neighbor cells influences when a cell divides. A collection of software and scripts existed to investigate this situation, but again was written in multiple languages, including the proprietary language IDL, primarily used in astronomy. The Schmidt Academy rewrote the IDL and C++ code into a single, easy to use python package that runs on Mac, Windows and Linux computers. An example of the recovered strain is shown below.
As a result of the collaboration, the Stathopoulos Lab now has a workspace on GitHub where all the lab software has a common home, and software best practices are better understood and used within the lab.